Josh
L. Morgan
 |
Josh
L. Morgan
|
 |
| Home |
CV |
Gallery |
Lichtman
Lab |
 |
Hi I am a neuroscientist specializing in imaging the development and synaptic organization of visual circuits. Current position: Postdoctoral fellow in the lab of Jeff W. Lichtman at Harvard University Future plans: I hope to continue my research into the organization of synaptic networks and I am in the process of applying for faculty positions. |
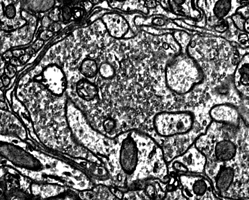
Why Connectomics? What do biological neural networks look like? It has been more than a hundred years since Ramon y Cajal figured out that cells in the brain were linked together by synapses and more than 70 years since the first computational models of neural networks were developed. Yet in the subsequent years of research it is rare that neuroscientists have been able to directly observe the network organization of neural circuits. Instead, canonical circuits have been constructed by studying one or two neurons at a time in many different subjects and combining these observations together into a composite circuit. While this approach is effective at capturing the most stereotyped features of neural circuits, they fail to capture the interdependency of neurons in real biological circuits. In particular, network properties that emerge only after extended periods of postnatal activity dependent development and learning are unlikely to be successfully inferred by looking at only one or two cells at a time. By imaging every cell and every synapse in a circuit at high resolution, large scale electron microscopy provides an unprecedented view of the synaptic organization of neural circuits. As reconstructing large networks with electron microscopy becomes easier the inferred organization of the nervous system can be replaced with direct observation of the interconnections of hundreds or thousands of neurons. Furthermore, neural networks mapped with electron microscopy reveal much more than which neurons are connected to one another. Large scale electron microscopy also reveals the number, position, size and ultrastructure of each synapse connecting every pair of neurons in a network.